Vista Browser Genome
.Part of thebook series (MIMB, volume 395) SummaryThis chapter discusses VISTA Browser and associated computational tools for analysis and visual exploration of genomic alignments. The availability of massive amounts of genomic data produced by sequencing centers stimulated active development of computational tools for analyzing sequences and complete genomes, including tools for comparative analysis. Among algorithmic and computational challenges of such analysis, i.e., efficient and fast alignment, decoding of evolutionary history, the search for functional elements in genomes, and others, visualization of comparative results is of great importance. Only interactive viewing and manipulation of data allow for its in-depth investigation by biologists.We describe the rich capabilities of the interactive VISTA Browser with its extensions and modifications, and provide examples of the examination of alignments of DNA sequences and whole genomes, both eukaryotic and microbial. VISTA portal ( ) provides access to all these tools. AcknowledgmentsThe author is grateful to Michael Cipriano and Alexander Levin for their help with the manuscript. The VISTA project is an ongoing collaborative effort of a large group of scientists and engineers.
It has been developed and maintained in the Genomics Division of Lawrence Berkeley National Laboratory. The names of all contributors are found at the VISTA website ( ).The project was partially supported by the grant no. HL88728, Berkeley-PGA, under the Programs for Genomic Application, funded by the US National Heart, Lung, and Blood Institute, and performed under Department of Energy Contract DE-AC0378SF00098, University of California.
AbstractGenome browser provides a graphical interface for users to browse, search, retrieve and analyze genomic sequence and annotation data. Web-based genome browsers can be classified into general genome browsers with multiple species and species-specific genome browsers. In this review, we attempt to give an overview for the main functions and features of web-based genome browsers, covering data visualization, retrieval, analysis and customization. To give a brief introduction to the multiple-species genome browser, we describe the user interface and main functions of the Ensembl and UCSC genome browsers using the human alpha-globin gene cluster as an example. We further use the MSU and the Rice-Map genome browsers to show some special features of species-specific genome browser, taking a rice transcription factor gene OsSPL14 as an example.
, INTRODUCTIONThe initial sequence generated by the human genome project together with the draft genome sequence of several model organisms, including the house mouse ( Mus musculus), fruit fly ( Drosophila melanogaster), nematode ( Caenorhabditis elegan), baker’s yeast ( Saccharomyces cerevisiae), Gram-negative bacterium ( Escherichia coli) and thale cress ( Arabidopsis thaliana), completed at the beginning of this millennium create a paradigm shift within biological research, as predicted by Gilbert in the early 1990s. With the rapid development of next-generation sequencing technologies, hundreds of eukaryotic and thousands of prokaryotic genomes have been sequenced. All the sequence data as well as the annotations generated through most completed or ongoing genome projects are collected in the genome databases and are publicly available through web portals such as the NCBI genome portal and the EBI genome database website. Great efforts were made in the construction of genome databases such as the human genome database (GDB) and the E. Coli genome database (Colibri) in the early 1990s to enable subsequent quantities of data to be accommodated.By systematic integration of genome sequences together with annotations generated through much heterogeneous data, genome browser provides a unique platform for molecular biologists to browse, search, retrieve and analyze these genomic data efficiently and conveniently. With a graphical interface, genome browser helps users to extract and summarize information intuitively from huge amount of raw data.
Furthermore, different types of annotations from multiple sources can be integrated into one genome browser, helping users to analyze data across different data providers effectively. With a uniform interface, users can navigate the whole genome using the same genomics coordinate system, and make comparative analysis across different lineages such as primates, mammalians, vertebrates and plants.In general, genome browser can be divided into web-based browsers and stand-alone applications.
Vista Promoter
In this review, we focus on web-based genome browsers which are useful in promoting biological research due to their data quality, flexible accessibility and high performance. First, dedicated organizations often collect and integrate high-quality annotation data into web-based genome browsers, providing plentiful up-to-date information for the community. Second, users can access them anywhere with a standard web browser, avoiding any additional effort of setting up local environment for application installation and data preparation.
Third, web-based genome browsers are usually installed on high performance servers and can support more complex and larger scale data types and applications. Here, we attempt to give a brief introduction to the main web-based genome browsers and the underlying frameworks. Web-based genome browsersCurrently, there are two types of web-based genome browsers.
The first type is the multiple-species genome browsers implemented in, among others, the UCSC genome database , the Ensembl project , the NCBI Map viewer website , the Phytozome and Gramene platforms. These genome browsers integrate sequence and annotations for dozens of organisms and further promote cross-species comparative analysis. Most of them contain abundant annotations, covering gene model, transcript evidence, expression profiles, regulatory data, genomic conversation, etc. Each set of pre-computed annotation data is called a track in genome browsers. The essence of a genome browser is to pile up multiple tracks under the same genomic coordinate along the Y-axis, thus users could easily examine the consistency or difference of the annotation data and make their judgments of the functions or other features of the genomic region. Lists several mainstream web-based genome browsers, including Ensembl, the UCSC genome browser and the NCBI Map Viewer, which are accessed by a large number of users worldwide.
NameURLDescriptionEnsemblMajor species with completed genome sequences providing lineage-specific web portals for vertebrates, metazoa, plants, fungi, protists and bacteria.UCSCMajor species with completed genome sequences including vertebrates, deuterostomes, insects and nematodes. NameURLDescriptionEnsemblMajor species with completed genome sequences providing lineage-specific web portals for vertebrates, metazoa, plants, fungi, protists and bacteria.UCSCMajor species with completed genome sequences including vertebrates, deuterostomes, insects and nematodes. NameURLDescriptionEnsemblMajor species with completed genome sequences providing lineage-specific web portals for vertebrates, metazoa, plants, fungi, protists and bacteria.UCSCMajor species with completed genome sequences including vertebrates, deuterostomes, insects and nematodes.
NameURLDescriptionEnsemblMajor species with completed genome sequences providing lineage-specific web portals for vertebrates, metazoa, plants, fungi, protists and bacteria.UCSCMajor species with completed genome sequences including vertebrates, deuterostomes, insects and nematodes. The other type is the species-specific genome browsers which mainly focus on one model organism and may have more annotations for a particular species. Powered by the Generic Model Organism Database (GMOD) project , dozens of open-source software tools are collected for creating and managing genome biological databases, and the GBrowse framework is one of the most popular tools in the GMOD project. Currently, most of these species-specific genome browsers are implemented based on the GBrowse framework, such as MGI, FlyBase, WormBase, SGD and TAIR. Genome browser frameworksThe creation of large sequencing data for various species increases the demand for building genome browsers to help researchers view and analyze these data in a more intuitive way. However, building a web-based genome browser from scratch is both time and labor consuming, while well-designed genome browser frameworks could be useful in this aspect.
As listed in, there are some web-based genome browser frameworks for users to install locally, and to configure and customize their own annotation data efficiently. In addition to providing rich resources, Ensembl and the UCSC systems are also released as software packages for local installation. GBrowse is the most popular genome browser framework and has been widely used in model organism projects for data visualization. The new genome browsers, JBrowse , ABrowse and Anno-J , support Google-map like navigation, and LookSeq is designed for raw sequencing reads presentation. Furthermore, there are some synteny genome browsers for comparative visualization of several species, with separated genome coordinates. The underlying technology of genome browsersTraditional genome browsers often employ classical web technologies based on synchronized transferring and static web pages.
By using the AJAX-based web technology, modern genome browsers could bring better user experience to scientists. Currently, most traditional genome browsers have actively employed this new web technology, but they still cannot provide as interactive browsing experience as the newly designed ones. For example, GBrowse supports smooth browsing within a limited region by preloading larger pictures (version 2.20 onwards) , while new AJAX-based browsers such as ABrowse , Anno-J and JBrowse support smooth dragging along the whole genome.Genome browser can be divided into two categories based on whether the image is rendered on the server side or on the client side.
Server-side rendering browsers such as UCSC, Ensembl, GBrowse and ABrowse extract the requested data from the back-end databases and render them into pictures on the server, and then send the pictures to the client web browsers. Client-side rendering browsers such as Anno-J and JBrowse send the requested data to client web browsers directly and draw the pictures dynamically in client web browsers. Client-side rendering reduces the server burden and the network data flow by distributing computing tasks to client sides.
On the other hand, the pictures produced by server side could bring users much richer details of the annotation data since current web browsers support limited drawing functions. FUNCTIONATILIES AND FEATURES VisualizationTaking the advantage of high-throughput sequencing technology and high-performance computing resource, immense volumes of genomic annotation data are being made available.
The principal function of the genome browser is to aggregate different types of annotation data together and integrate them into an abstract graphical view. Annotations are organized under a uniform genome coordinate with the chromosome as the X-axis and various types of data being displayed along the Y-axis.A web-based genome browser often provides a centralized or a set of databases to store different types of annotation data obtained from several organizations. The challenge for the general genome browsers is how to display this information properly for different genomic scales. Massive amounts of information need to be incorporated into the picture when a large genomic area is requested, which could overburden the server and the network. And too many heavy and complicated details also disturb the user attention. The UCSC genome browser tries to solve the problem by providing multiple views for a track.
The dense view of some tracks could be displayed to hide complicated details when zooming out to a large area of a chromosome, so that the user has a broad picture of the selected chromosome region.It is useful to have an overview of a large area of the chromosome and look into several small regions for details simultaneously. Putting paralogous genes together in one page could promote comparative analysis greatly. NCBI sequence viewer supports users to view different regions inside the same chromosome, providing flexible multiple-panel-based navigating approach with different color cursors indicating the corresponding genomic locations. ABrowse supports users to visualize multiple genome regions of different chromosomes/genomes in separated in-page windows , while the separated windows are not fully operable as the main browsing canvas.Displaying genome alignments within or between species helps users to make comparative studies such as finding the conserved or fast involved elements among several genomes.
Examples of this type of browsers are the Generic Synteny Browser , Sybil , SynBrowse , SynView and VISTA. Although some of the general genome browsers could also display genomes alignments, the alignments could only be organized under the coordinates of one of the genomes. Most of the synteny browsers could do it in a better way by arranging each segment of the alignment under the coordinate of its own genome while piling them together.
This type of genome browsers often focus on sequence alignment of DNA or protein sequences rather than other types of annotation data.The NGS raw data viewer aims to provide graphic views for the short read sequences aligned to the genome and helps users to find genome structure variations. LookSeq provides a simple graphical representation for paired sequence reads, which helps to reveal potential insertions and deletions. Users can have a view of high-depth original reads besides general annotation results, and manually manipulate them to assimilate information at different levels of resolution.
Data retrieval and analysisIn addition to graphical data navigation, data retrieval and analysis are useful features for a genome browser. Most of the existing genome browsers support search functions to locate genomic regions by coordinates, sequences or keywords. Some genome browsers employ a system to retrieve bulk data. For example, the UCSC system offers Table Browser to retrieve specified datasets , while the Ensembl, Gramene and ABrowse projects employ the BioMart system for making large data queries.To facilitate further data analysis, multiple data access approaches are supported for analysis tools to retrieve data from the genome browsers. The Galaxy genome browser Trackster supports analysis by integrating tools in the same platform, connecting data manipulation with visualization tightly.
The users can view the data in the genome browser seamlessly and further filter the visualized data on-the-fly, which helps to refine the results conveniently and efficiently. Some genome browsers offer plug-in mechanism, supporting third-party development and integration of tools based on the open interface, so that various tools could be added and launched easily within one platform.
Furthermore, it is also valuable for genome browsers to have close integration with external applications to perform the sequence and annotation data analysis transparently. The UCSC genome browser supports users to submit selected data directly to Galaxy and GREAT platforms, while ABrowse supports data submission to Galaxy and WebLab for data analysis.In addition to human-oriented interfaces, machine-oriented data retrieval is becoming even more essential for large-scale data analysis. Currently, web service has been widely used for exchanging structured information through networks among various data resources. ABrowse supports native standard SOAP-based web service for underlying data access , which is also supported in BioMart and employed by the Ensembl and Gramene projects. In addition, BioDAS has been widely used in some genome browsers for data exchange of distributed platforms. CustomizationIt is much easier to build a genome browser based on a framework. Most of the frameworks have configuration files for users to customize local data.
Currently, it is easy for users to integrate annotation into general genome browsers with several popular data formats, such as GFF, BED, SAM and WIG. However, if the data format is not compatible with the genome browser, it is difficult for average users to convert data formats to meet the system requirements. Some browser frameworks such as GBrowse and ABrowse provide plug-in mechanisms or API to extend new data types ,.The genome browser is fast becoming a collaboration platform for researchers to share discoveries and to exchange knowledge , promoting remote cooperation among a group of scientists. Most genome browsers provide a facility for end-users to upload, create and share their own annotation data, providing a collaborative platform. The user annotation comments can also be attached for selected items on-the-fly and shared with specified users and groups , or the whole research community.
Besides, users can save any important analysis status as bookmarks or sessions , and share them efficiently among several researchers.To summarize, we list the main functions including visualization, data retrieval and analysis, and customization for different genome browsers in. Currently, there are many applications built on these platforms. For example, the Epigenome Roadmap Genome Browser and Cistrome are built based on the UCSC system; Gramene is built based on Ensembl; Rice-Map is built based on ABrowse; Flybase and Wormbase are built based on GBrowse; and the Arabidopsis epigenome map is built based on Anno-J. These applications provide valuable resources for biologists with the support of general genome browser platforms.
EXAMPLESAs described above, genome browser is a useful tool in computational molecular biology, especially in genetic, genomic and evolutionary researches. In the following section, we introduce the main functions of general genome browsers using the Ensembl and UCSC genome browsers as examples. We further describe the features of species-specific genome browsers based on the MSU and Rice-Map genome browsers. General genome browsersHemoglobin is the key molecule used to transport oxygen in vertebrates.
Human adult hemoglobins are encoded by the alpha-globin and beta-globin gene clusters. Taking the human alpha-globin gene cluster as an example, we describe the features of the Ensembl and UCSC genome browsers. The screenshot of the user interface of the Ensembl and UCSC genome browsers for the alpha-globin gene cluster located in the forward strand of chromosome 16, consisting of five members (HBZ, HBM, HBA2, HBA1 and HBQ1) and two pseudogenes.
( a) The user interface of the Ensembl genome browser with default setting of the annotation tracks, showing the alpha-globin gene cluster. The graphical annotations are displayed in the main body divided in three sections from top to bottom. The top section shows the location of this alpha-globin gene cluser on chromosome 16, where the selected region is highlighted by a red box. The middle section shows the chromosome band, the contigs and the protein-coding genes annotated by the Ensembl/Havana project. The bottom section shows more annotations in this 35 kb region. ( b) The main user interface of the UCSC genome browser showing the default tracks in default order for the human alpha-globin gene cluster. The top graphical window shows the annotations, and the text window below supports users to make configuration simultaneously in the same page.
User can choose four styles to view different tracks, i.e. The dense, squish, pack and full styles. Some of the groups and tracks are similar to that of Ensembl, e.g. The annotations for genes, mRNAs and ESTs, while some are different, e.g. The group of phenotype and disease associations. The screenshot of the user interface of the Ensembl and UCSC genome browsers for the alpha-globin gene cluster located in the forward strand of chromosome 16, consisting of five members (HBZ, HBM, HBA2, HBA1 and HBQ1) and two pseudogenes.
( a) The user interface of the Ensembl genome browser with default setting of the annotation tracks, showing the alpha-globin gene cluster. The graphical annotations are displayed in the main body divided in three sections from top to bottom. The top section shows the location of this alpha-globin gene cluser on chromosome 16, where the selected region is highlighted by a red box. The middle section shows the chromosome band, the contigs and the protein-coding genes annotated by the Ensembl/Havana project. The bottom section shows more annotations in this 35 kb region. ( b) The main user interface of the UCSC genome browser showing the default tracks in default order for the human alpha-globin gene cluster. Zulu camp at shambala game reserve. The top graphical window shows the annotations, and the text window below supports users to make configuration simultaneously in the same page.
User can choose four styles to view different tracks, i.e. The dense, squish, pack and full styles. Some of the groups and tracks are similar to that of Ensembl, e.g. The annotations for genes, mRNAs and ESTs, while some are different, e.g.
The group of phenotype and disease associations. Data visualizationThe Ensembl genome browser (Release 67) provides the same user interface for each organism. By choosing ‘Human’ as the target organism and searching the keyword ‘alpha hemoglobin’, then following the links in the search results page, users can enter the chromosome region 16:226,679-227,521 where the HBA1 gene encoding alpha hemoglobin is located. By expanding the region to chromosome 16:200,001-235,000, user can have a view of the alpha-globin gene cluster with default setting (a). The main body of the interface contains two panels. The left panel lists the main menu for location-based displays at different levels from whole genome, chromosome summary to region overview and region in detail.
And links to comparative genomics, genetic variation as well as sequence markers are also provided. The main panel is arranged in three sections from top to bottom, providing different scales for users to analyze the genome.In addition to the location view, Ensembl provides separate pages to display various types of information, organized in a tabbed structure. In the gene page, different transcripts for HBA1 are organized in a table, and the summary of this gene is described below, with related transcripts highlighted in light green. Furthermore, users can jump to the transcript page by clicking the individual transcript.
The detailed transcript descriptions are shown in the middle panel. In addition, other information such as regulation and variation can also be displayed in different pages for users to investigate with different views, and cross-links are also listed in the left panel for specific pages. Detailed description of the user interface can be found in the Ensembl online tutorial.Besides the general functionalities as described above, the main panel has some special features designed for end-users to access the annotations conveniently. Clicking on the graphical item in the main panel will show the detailed information of this annotation unit.
Moving the mouse over the track name at the left side of each track will show several logos. Detailed track description will be shown by moving the mouse over the information logo, and the display style of each track can be changed by moving the mouse to the configuration logo. Furthermore, users can click and drag the column bar beside the track name to re-order the track location freely for customized visualization.The interface of the UCSC genome browser (v267, hg19) has some common features with that of the Ensembl genome browser, including the overall layout for graphical display of chromosome coordinates, the search boxes to make location or text queries and the layout of exon–intron structure of the genes. However, there are some special features implemented by the UCSC genome browser (b).The default tracks displayed are different from that of Ensembl.
The annotation of protein-coding genes and their transcripts include the UCSC genes, the RefSeq genes, the Human mRNAs and the spliced ESTs. For example, the UCSC and RefSeq annotation for the human alpha-globin gene cluster including HBZ, HBM, HBA2, HBA1 and HBQ1 are displayed together with their mRNA and spliced EST transcript evidence. Besides, the histone modification H3K27Ac mark on seven cell lines, the digital DNaseI hypersensitivity clusters and the transcription factor ChIP-Seq assay generated by the ENCODE project are also displayed as default tracks. The blue peaks show that all these genes have H3K27Ac histone mark enrichment in the K562 cell line, while the two red peaks under the HBZ and HBM genes show the enrichment in the GM12878 cell line.
Furthermore, multiple alignments of 46 vertebrate species are provided to measure the evolutionary conservation of this region. And most genes have high conservation value among mammals, while the intergenic regions are less conserved. SNP variants from dbSNP, repeating elements generated by RepeatMasker are also provided as default tracks.The user experience of UCSC genome browser is different from that of the Ensembl system. Users can freely drag the graphical canvas upstream or downstream, and right-click functions are provided for the whole canvas for quick configuration. In contrast to the Ensembl system, the column bar is linked to track description and configuration page, and the tracks can be re-ordered by clicking and dragging the track name.
Data retrieval and analysisIn addition to data browsing, data analysis is also supported by the Ensembl platform. A BioMart-powered query system is provided for bulk data retrieval, and advanced users can use standard Mart API to retrieve bulk data from the backend database directly. Furthermore, sequence similarity searches with BLAST/BLAT and other tools such as assembly converter, ID history converter, region report and variant effect predictor are supported for users to process data conveniently.As for the UCSC system, some other useful analysis functions are also supported besides data navigation. Different from the BioMart retrieval system employed by the Ensembl platform, the UCSC platform offers the Table Browser system for bulk data retrieval, supporting operations of intersection and correlation for dataset processing. And the retrieved result can be directly sent to external platforms for further analysis with a single click. In addition to the sequence similarity search using BLAT, the UCSC system provides some online tools for users to perform quick data analysis, such as In Silico PCR, Gene Sorter, VisiGene and several handy utilities. CustomizationIn the left panel of the Ensembl browser page, three buttons ‘Configue this page’, ‘Manage your data’ and ‘Export data’ for customization can be invoked to add or remove annotation tracks, and to upload and analyze users’ own data.
Users can also add annotations freely for specified items as user comments. To support quick link to previous browsing status, users can bookmark the layout of the current browser for future study. To promote knowledge sharing, group mechanism is supported in the Ensembl system for collaboration among several colleagues.To facilitate data customization, the UCSC system allows users to save settings as sessions for restoring and sharing through a wiki system. And users can also upload tracks for personal data visualization and analysis with the precomputed annotations. Moreover, the UCSC system provides the Track Hubs functionality on the home page, offering a sharing mechanism for large custom datasets from other individuals and labs. Species-specific genome browsersAs listed in, dozens of species-specific genome browsers are available online.
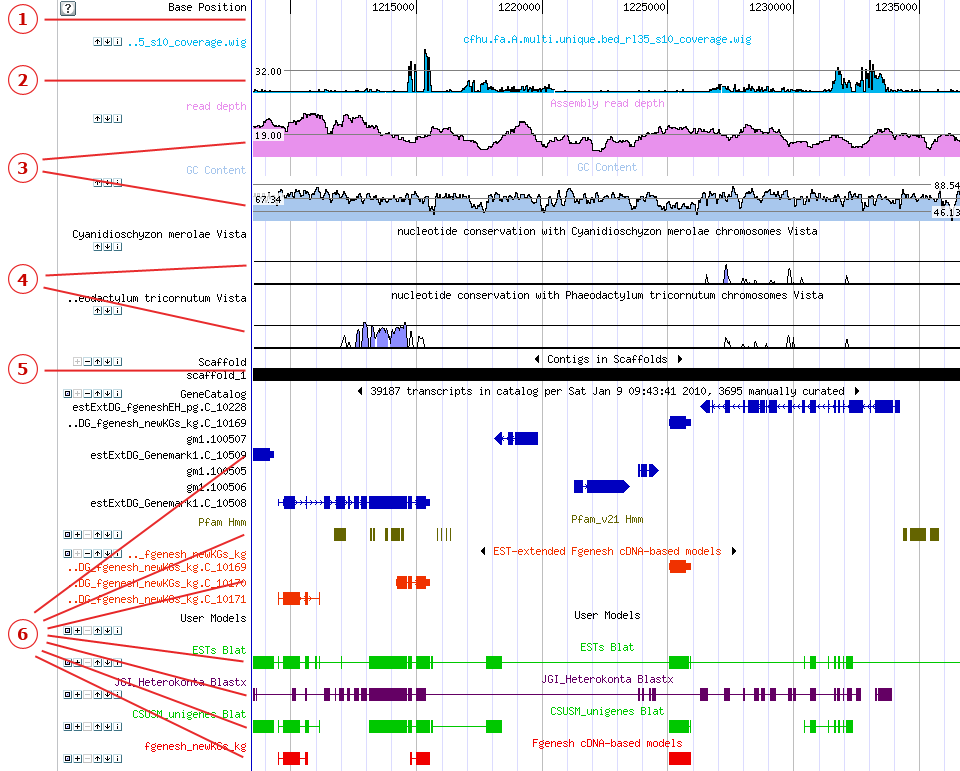
Here, we introduce the two species-specific genome browsers dedicated to the rice genome, the MSU rice genome browser and the Rice-Map genome browser using the rice transcription factor OsSPL14 as an example. Recently, OsSPL14 has been reported as a member of the rice SBP-like gene family and is essential for rice grain productivity. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice, promoting panicle branching and enhanced grain yield ,. The screenshot of the user interface of the MSU and Rice-Map genome browsers for the rice transcription factor OsSPL14. ( a) The user interface of the MSU rice genome browser. The chromosome overview is displayed at the top, the regional view is shown at the middle and the bottom section is the detailed view for four annotation tracks including gene model, expression data for pre-emergence inflorescence, pistil and embryo—25 days after pollination stages. ( b) The Rice-Map genome browser.
In the middle canvas, different annotation tracks are listed, including the MSU gene model, and three comparative genome alignments from VISTA. The detailed information for individual entries is shown in the right panel, interpreting the data resource, entry location, sequence and function etc. The screenshot of the user interface of the MSU and Rice-Map genome browsers for the rice transcription factor OsSPL14. ( a) The user interface of the MSU rice genome browser. The chromosome overview is displayed at the top, the regional view is shown at the middle and the bottom section is the detailed view for four annotation tracks including gene model, expression data for pre-emergence inflorescence, pistil and embryo—25 days after pollination stages.
( b) The Rice-Map genome browser. In the middle canvas, different annotation tracks are listed, including the MSU gene model, and three comparative genome alignments from VISTA. The detailed information for individual entries is shown in the right panel, interpreting the data resource, entry location, sequence and function etc.
Data visualizationIn the MSU rice genome browser (Release 7), users can search the OsSPL14 gene by specifying ‘SPL14’ in the search box. As shown in a, OsSPL14 is encoded by the transcript LOCOs08g39890.1 with three exons, and located in the reverse strand of chromosome 8, from 25 274 449 to 25 278 696. Powered by the GBrowse platform, the MSU rice genome browser provides annotation views with different scales, including chromosome overview, regional view and detailed view. The large-scale view provides a broader picture for users to inspect the upstream and downstream annotation conveniently. In the detailed annotation canvas, more than 82 annotation tracks are provided, covering gene model, transcript evidence, expression profiling, sequence alignment, genetic marker, SNP, RNA-Seq coverage and other genomic features. In addition to the basic gene model information, users may inspect this gene in different development stages through various RNA-Seq expression data. According to the integrated expression data, this gene has a highly expressed signal in the pre-emergence inflourescence stage, pistil stage and embryo-25 days after pollination stage, which is consistent with SBP-box gene function reported in the literature (a).In the Rice-Map genome browser (v1.0), different annotation tracks are organized in a map-like visualization canvas, with the name of opened tracks listed in the right panel.
Currently, 81 japonica tracks and 82 indica tracks have been compiled and loaded into Rice-Map. Besides basic gene annotation, there are rich annotation for cross genome alignments and conservation values, offering important clues to investigate this gene in other plants. From the conservation data, users can find that the sequence of this gene is highly conserved among rice and other grasses including the wild grass ( Brachypodium distachyon), maize ( Zea mays) and sorghum ( Sorghum bicolor), especially in the exon regions (b). Furthermore, users can use the ‘magic wand’ in the navigation panel (left-top corner) to select an interesting region by mouse, and inspect it in the main canvas or in a new sub-window.
After clicking the annotation item, detailed descriptions for track and entry are provided in the right panel, along with the graphical view. Data retrieval and analysisThe sequence and annotation data can be dumped from the MSU genome browser database through the online user interface. In the Rice-Map platform, a BioMart-powered system ‘Rice Mart’ is provided for users to retrieve large datasets, with a standard interface. The retrieved dataset, as well as the genomic, CDS and protein sequence information of each entry can also be submitted to external bioinformatic platforms, i.e. WebLab and Galaxy for further analysis. CustomizationThe MSU genome browser supports users wishing to view their own data in the genome browser, alongside the precomputed annotations.
By uploading the local data file or specifying the URL of a remote annotation file, users can easily integrate their own data into the online platform.In the Rice-Map platform, users can add user annotation by clicking and dragging a genomic region freely, providing a more flexible approach for average users to add annotation on-the-fly. Furthermore, Rice-Map provides a multi-functional user space for users to save contributed data online, including track evaluation, entry comments and browsing landmarks.
Moreover, users can also set privilege for the contributed items, and the private items can be taken as personal online notebook, while the public items can be shared among the whole community. CHALLENGES AND PERSPECTIVESThe web-based genome browsers have many challenges such as how to visualize different data types produced by the NGS sequencing instruments. For the annotation visualization, FlyBase tries to provide a 3D view for different development stages of RNA expression data in one track, providing an overall view of differential gene expression patterns over and above the traditionally separated tracks. As for the circular genomes, presenting them as circular maps is more natural than linear views, and Genome Projector provides new circular views for more than 400 bacterial genomes. For the original reads presentation, LookSeq can provide a reads view for small genomes currently but performs poorly with large-scale genomes.
Based on the HTML5 language, semantic tags and web storage are introduced into the client browser, which helps users to improve their browsing experience efficiently. With the launch of the large-scale sequencing projects such as 1000 Genomes, visualizing individual sequencing data becomes a big challenge. It is necessary to develop new approaches to visualize several individual genomes together under a coordinate-free system to figure out insertions and deletions conveniently.Since personalized functions are becoming more and more crucial to promote scientific research, some novel features are being added to genome browsers, improving user participation, collaboration and communication.
The WebApollo project is now aiming to develop an online collaborative annotation editor, allowing users to interactively create and edit genomic annotations in a web-based graphical environment. The management and transmission of large-scale data are becoming a great challenge. Some genome browsers set the data storage lifespan and limit the data size for custom tracks ,. High-speed network facilities and powerful servers are needed to solve such problems in the future.
Furthermore, a distributed system is needed to exchange data across different platforms and resources to achieve a collaborative society for biologists , such as the P2P data transmission approach, enabling user participation in data sharing.Though web service is a becoming a standard protocol for data exchange and application communication, the problem of how to define the data exchange format and the application interface is still unsolved. Putting the genome browser and the bioinformatics application platform on a cloud environment and allowing them to share the same storage could be a possible solution to avoid heavy data transmission. Due to the demand of access speed and large-scale data integration, web-based genome browsers are gradually moving to cluster servers or cloud environments. UCSC provides powerful hardware to offer a high-speed browsing experience , while Ensembl and JBrowse are actively using Amazon web services to improve the online service ,.
In the future, more and more cloud technologies would provide high performance for the end users.In addition, genome browsers may take advantage of the latest hardware technology to offer a richer presentation of data. For example, GenomePad is a novel mobile-based genome browser, supporting users to navigate and share genome information conveniently on mobile phones.
Genome wowser is an iPad-enabled human genome browser, providing an intuitive and portable presentation of the popular UCSC genome browser. Moreover, JBrowse is also planning to use mobile devices for sequencing application.